Chip Seq Data Analysis Workshop On April 12 Easeq Interactive Chip This easeq tutorial will show you: 1) how to visualize chip seq signal at all genes in the genome sorted according to their sizes, and 2) how to gate out the shortest genes. How to make heatmaps of chip seq signal at genes and sort them according to sizes. how to make ratiometric heatmaps, calculate fold difference, and sort the regions accordingly.
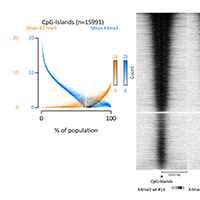
Data Easeq Interactive Chip Seq Analysis Software How to import chip seq data and a set of genomic regions into easeq (the example data can be downloaded here). how to make heatmaps from the imported data above. in the example we visualize histone marks (h3k27me3 and h3k36me3) at cpg islands and sort them according to their sizes. how to access the autogenerated legends and methods. This includes making basic plots; finding, annotating, sorting, and filtering of peaks; using easeq as a genome browser; measuring chip seq signal and calculating ratios; as well as data import and export. In this tutorial we will: look at distribution of enriched regions across genes. datasets for this tutorial were generated in the lab of frank pugh. for this analysis, we will be using chip exo datasets. for this experiment, immunoprecipitation was performed with antibodies against reb1. In this tutorial, we will work through the chip seq dataset. check the methods section in the paper for more details on the chip seq library preparation, following the standard procedure. 1. download fastq files directly from ena website.
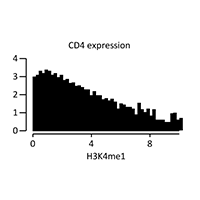
Data Easeq Interactive Chip Seq Analysis Software In this tutorial we will: look at distribution of enriched regions across genes. datasets for this tutorial were generated in the lab of frank pugh. for this analysis, we will be using chip exo datasets. for this experiment, immunoprecipitation was performed with antibodies against reb1. In this tutorial, we will work through the chip seq dataset. check the methods section in the paper for more details on the chip seq library preparation, following the standard procedure. 1. download fastq files directly from ena website. In this tutorial, we will try to give step by step examples exercises on how to use our chip seq methods to analyse transcription factor binding sites, histone modifications, and cage data. Download the data used for the video tutorial video: how to visualize chip seq signal at all genes in the genome sorted according to sizes how to gate out the shortest genes. A complete workflow for the analysis of full size chip seq (and similar) data sets using peak motifs. nat protoc. 7:1551 68. g. (2010) great improves functional interpretation of cis regulatory regions. nat biotechnol. 28:495 501. This easeq tutorial will show you how to: 1) quantify chip seq signal at transcription start sites, 2) isolate genes with a high level of h3k27me3, and 3) sort them according to log2 fold.
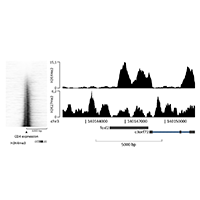
Data Easeq Interactive Chip Seq Analysis Software In this tutorial, we will try to give step by step examples exercises on how to use our chip seq methods to analyse transcription factor binding sites, histone modifications, and cage data. Download the data used for the video tutorial video: how to visualize chip seq signal at all genes in the genome sorted according to sizes how to gate out the shortest genes. A complete workflow for the analysis of full size chip seq (and similar) data sets using peak motifs. nat protoc. 7:1551 68. g. (2010) great improves functional interpretation of cis regulatory regions. nat biotechnol. 28:495 501. This easeq tutorial will show you how to: 1) quantify chip seq signal at transcription start sites, 2) isolate genes with a high level of h3k27me3, and 3) sort them according to log2 fold.
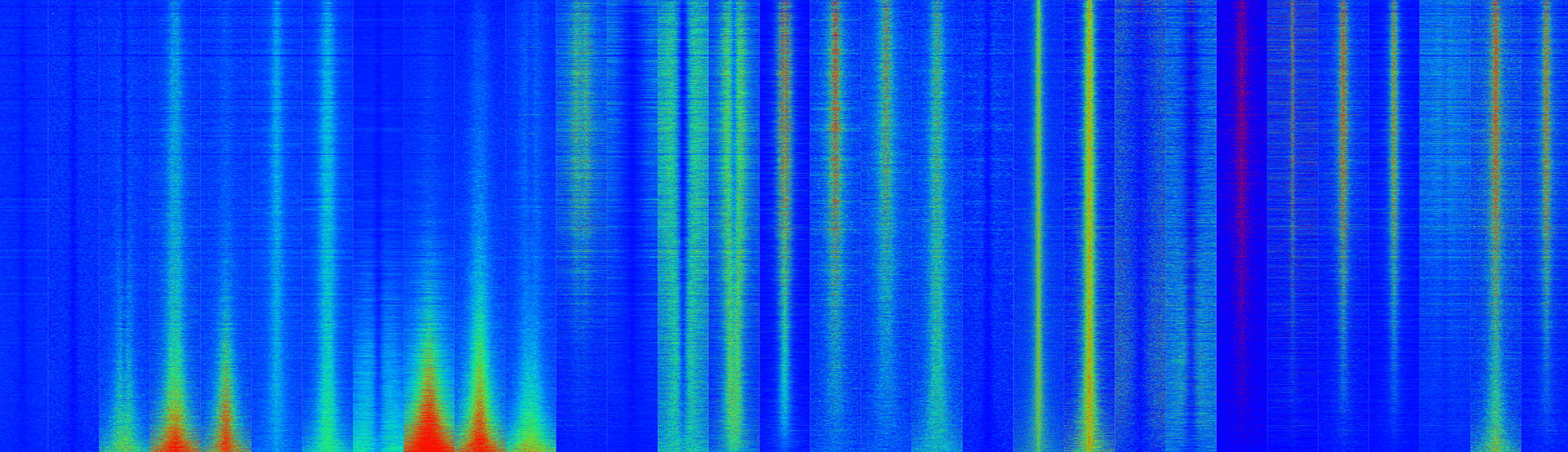
Data Easeq Interactive Chip Seq Analysis Software A complete workflow for the analysis of full size chip seq (and similar) data sets using peak motifs. nat protoc. 7:1551 68. g. (2010) great improves functional interpretation of cis regulatory regions. nat biotechnol. 28:495 501. This easeq tutorial will show you how to: 1) quantify chip seq signal at transcription start sites, 2) isolate genes with a high level of h3k27me3, and 3) sort them according to log2 fold.