
Impact Of Gene Annotation On Rna Seq Data Analysis Rna Seq Blog How to download gene annotation data and extract tsses as new regions. how to gate out regions and instruct new plots to show this population. how to handle plots and change their settings. easeq also contains a set of integrated tutorials that can guide you through how to apply a particular visualization or analysis to your own data. How to download a ‘geneset’ (annotation file) from ucsc and extract a regionset (a set of genomic loci) that correspond to the transcription start sites in this geneset using easeq.

Image Extractannotations Genomics Step2 How to use easeq as a genome browser and explore the underlying loci in a heatmap. how to download gene annotation data and extract tsses as new regions. how to gate out regions and instruct new plots to show this population. how to handle plots and change their settings. Subsets of regions can be gated into new sets (11). lists (12) and heatmaps (13) can be used to inspect individual loci ensuring that interpretations match underlying signals (14). examples in the videos here and here. Once you have an ensdb package for your genome, you can use the function getgrangesfromensdb in signac to extract a set of genomic ranges from the database for downstream use. see the signac vignette for an example. see here for tips for if your species does not have an annotation available on bioconductor marked as answer 1 1 reply. Either the mm10 coordinates for the tsses can then be imported as a regionset too, or the refseq [13] annotation for mm10 genes can be downloaded from ucsc [11, 12] and stored as a geneset within easeq (covered in subheading 3.1.4, steps 1 and 2).
Image Extractannotations Genomics Step3 Once you have an ensdb package for your genome, you can use the function getgrangesfromensdb in signac to extract a set of genomic ranges from the database for downstream use. see the signac vignette for an example. see here for tips for if your species does not have an annotation available on bioconductor marked as answer 1 1 reply. Either the mm10 coordinates for the tsses can then be imported as a regionset too, or the refseq [13] annotation for mm10 genes can be downloaded from ucsc [11, 12] and stored as a geneset within easeq (covered in subheading 3.1.4, steps 1 and 2). Extract transcript regions for bioinformatics on genome annotation sets. decompose a ucsc knowngenes file or ensembl derived gtf into transcript regions (i.e. exons, introns, utrs and cds). For genesets, easeq has an integrated menu to download the most current refseq annotation from ucsc. we recommend using this by default to get coordinates for gene features. a good starting point for finding regionsets would be to download it from the ucsc table browser or download e.g. a published peak set from geo. Either the mm10 coordi‐nates for the tsses can then be imported as a regionset too, or the refseq[13] annotation for mm10 genes can be downloaded from ucsc[12,11] and stored as a geneset within easeq (covered in 3.1.4 steps m‐n). Easeq free and interactive software for analysis and visualization of chip sequencing data news about learn videos.

Distribution Of The Annotated Tss Regions Gene Symbols Across 8 Extract transcript regions for bioinformatics on genome annotation sets. decompose a ucsc knowngenes file or ensembl derived gtf into transcript regions (i.e. exons, introns, utrs and cds). For genesets, easeq has an integrated menu to download the most current refseq annotation from ucsc. we recommend using this by default to get coordinates for gene features. a good starting point for finding regionsets would be to download it from the ucsc table browser or download e.g. a published peak set from geo. Either the mm10 coordi‐nates for the tsses can then be imported as a regionset too, or the refseq[13] annotation for mm10 genes can be downloaded from ucsc[12,11] and stored as a geneset within easeq (covered in 3.1.4 steps m‐n). Easeq free and interactive software for analysis and visualization of chip sequencing data news about learn videos.
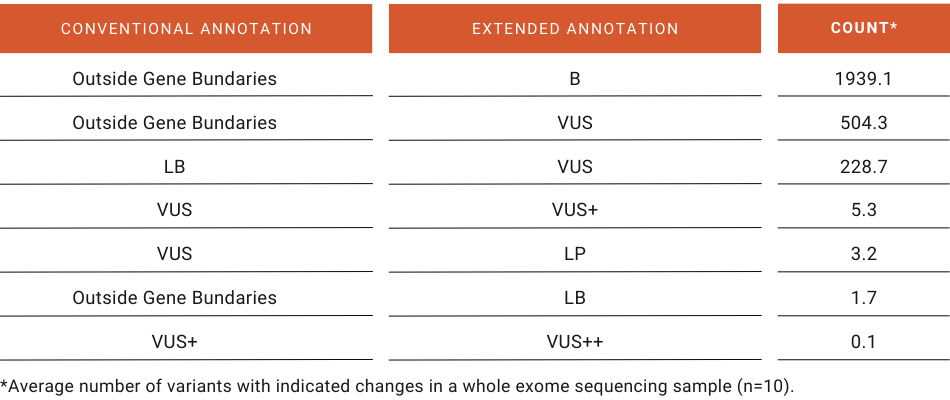
Seq S Extended Annotation A Solution For Alternative Isoforms In Ngs Either the mm10 coordi‐nates for the tsses can then be imported as a regionset too, or the refseq[13] annotation for mm10 genes can be downloaded from ucsc[12,11] and stored as a geneset within easeq (covered in 3.1.4 steps m‐n). Easeq free and interactive software for analysis and visualization of chip sequencing data news about learn videos.

International Hands On Workshop On Gene Annotation Data Train The