
Encode Pdf Human Genome Gene Expression The image shows selected encode and other gene and transcript annotations in the region of the human tp53 gene (region chr17:7,560,001–7,610,000 from the human february 2009 (grch37 hg19) genome assembly). The human epigenome has been experimentally characterized by thousands of measurements for every basepair in the human genome. we propose a deep neural network tensor factorization method, avocado, that compresses this epigenomic data into a dense, information rich representation.

Encode Encyclopedia Of Dna Elements Encode Data and derived results are made available through a freely accessible database. here we provide an overview of the project and the resources it is generating and illustrate the application of encode data to interpret the human genome. 1) the encyclopedia of dna elements (encode) project aims to identify all functional elements in the human genome through a worldwide consortium of research groups. 2) encode generates genomics data, tools, and methods through analyzing different cell types and assays to build a comprehensive database of functional elements. Encode data have revealed the regulatory impact of disease associated variants on gene expression, chromatin accessibility, and transcription factor binding, providing mechanistic insights into the genetic basis of disease susceptibility and pathogenesis. Encode data are now available for the entire human genome. all encode data are free and available for immediate use via : to search for encode data related to your area of interest and set up a browser view, use the ucsc experiment matrix or track search tool (advanced features).
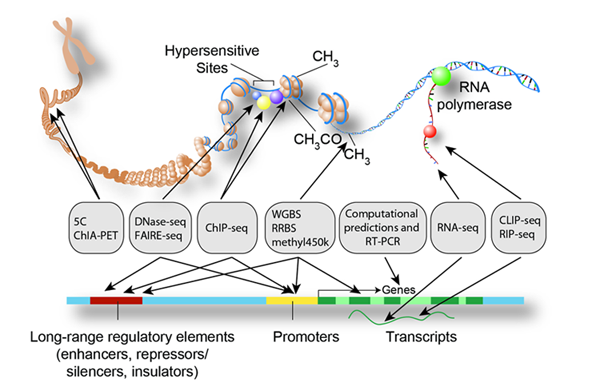
Encode Encyclopedia Of Dna Elements Encode Encode data have revealed the regulatory impact of disease associated variants on gene expression, chromatin accessibility, and transcription factor binding, providing mechanistic insights into the genetic basis of disease susceptibility and pathogenesis. Encode data are now available for the entire human genome. all encode data are free and available for immediate use via : to search for encode data related to your area of interest and set up a browser view, use the ucsc experiment matrix or track search tool (advanced features). On june 14, 2007, a report summarizing the findings of the pilot project revealed pervasive transcription of the human genome, with the majority of nucleotides represented in transcripts in at least a limited number of cell types at some time (the encode project consortium 2007). many of these transcripts comprised novel noncoding rna genes. Operationally, we define a functional element as a discrete genome segment that encodes a defined product (e.g., protein or non coding rna) or displays a reproducible biochemical signature (e.g., protein binding, or a specific chromatin structure). The gencode consortium aims to identify all gene features in the human genome using a combination of computa tional analysis, manual annotation, and experimental validation. Here we provide an overview of the project and the resources it is generating, and illustrate the application of encode data to interpret the human genome. interpreting the human genome sequence is one of the leading challenges of 21st century biology (collins et al., 2003).