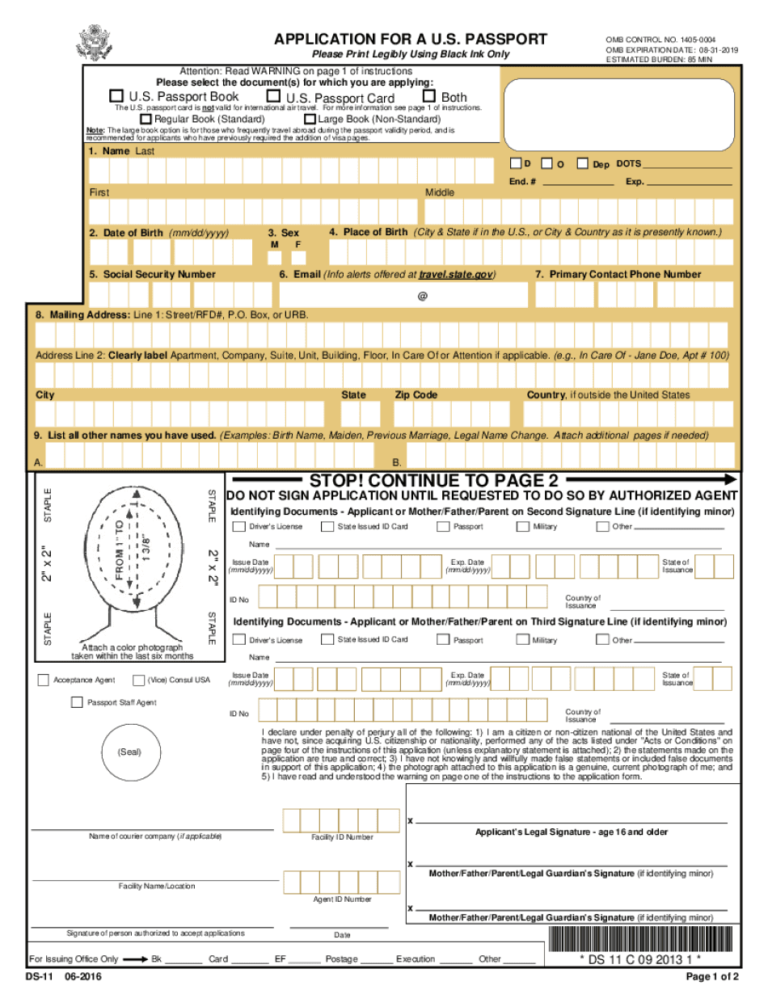
Ds 11 Form 2018 Fillable Printable Online Pdf Sample Can someone help with with setting up a calculation to run the new composite scheme pbeh 3c in gaussian? the code has been implemented in turbomole and orca and it's pretty easy to run it there. 3 i am conducting an organic synthesis involving stille coupling, using pdx2(dba)x3 p d x 2 (d b a) x 3 and p(o tol)x3 p (o t o l) x 3 as catalysts. reaction solvent is toluene in which the catalysts also dissolve. many texts said that celite pad can filtrate these catalysts, but i failed.

Us Passport Ds 11 Form Pdf Printable Form 2024 Question 3. there are some tailings. does existence of tailing always mean the band widening when i run the column chromatography? i heard that when i do the column, i had better use as minimum solvent as possible. but to prevent tailing, should i dilute the sample more? (i think the size of spot was small enough) question 4. This run was meant to be a sort of mock forensics experiment. there is dna from the "crime scene", "suspect 1", and "suspect 2". there are 3 samples from each, one is untreated, one is digested with ecorv, and one is digested with psti. Also today it is quite common to have the dna stain already in the gel while the electrophoresis is running (instead of adding a staining solution at the end of the run). this allows to follow the dna run in "real time". First! a buret reading should be to the estimated 0.01 ml, the first insignificant figure not to the gradations of 0.1 ml. one titration is one data point. if you want to check your overall precision the procedure is to run complete replicate samples on the one sample or [even on simultaneous replicate samples to check on the sampling technique]. since only 2 or 3 replicates are usually run.

Fillable Ds11 Form Printable Forms Free Online Also today it is quite common to have the dna stain already in the gel while the electrophoresis is running (instead of adding a staining solution at the end of the run). this allows to follow the dna run in "real time". First! a buret reading should be to the estimated 0.01 ml, the first insignificant figure not to the gradations of 0.1 ml. one titration is one data point. if you want to check your overall precision the procedure is to run complete replicate samples on the one sample or [even on simultaneous replicate samples to check on the sampling technique]. since only 2 or 3 replicates are usually run. Has anyone seen a comparative list of typical run times of the techniques used in analytical chemistry? i am making a list of rough timescales of analytical techniques (for a slide). assume that the. In gaussian, there is a stable keyword that checks the stability of the wavefunction. using stable=opt reoptimizes the wavefunction until a stable one is found if there is an instability. my questi. I am playing around with a prostaglandin molecule (23 heavy atoms) in gaussian09. my goal is to calculate the vertical ionization potential and electron affinity for the molecule. i already managed. Just few suggestions for finding a stationary point. you can add some additional angular flexibility into the basis by changing it to, say, 6 31g(2df,p) or even switch to more modern ahlrichs' def2 bases (def2svp to start from). besides, you'd better use ultrafine integration grid and tight optimization criteria and do geometry optimization and frequency calculation in a single run: opt=tight.

Fillable Online Ds11 Passport Form Pdf Ds11 Passport Form Pdf How Do Has anyone seen a comparative list of typical run times of the techniques used in analytical chemistry? i am making a list of rough timescales of analytical techniques (for a slide). assume that the. In gaussian, there is a stable keyword that checks the stability of the wavefunction. using stable=opt reoptimizes the wavefunction until a stable one is found if there is an instability. my questi. I am playing around with a prostaglandin molecule (23 heavy atoms) in gaussian09. my goal is to calculate the vertical ionization potential and electron affinity for the molecule. i already managed. Just few suggestions for finding a stationary point. you can add some additional angular flexibility into the basis by changing it to, say, 6 31g(2df,p) or even switch to more modern ahlrichs' def2 bases (def2svp to start from). besides, you'd better use ultrafine integration grid and tight optimization criteria and do geometry optimization and frequency calculation in a single run: opt=tight.
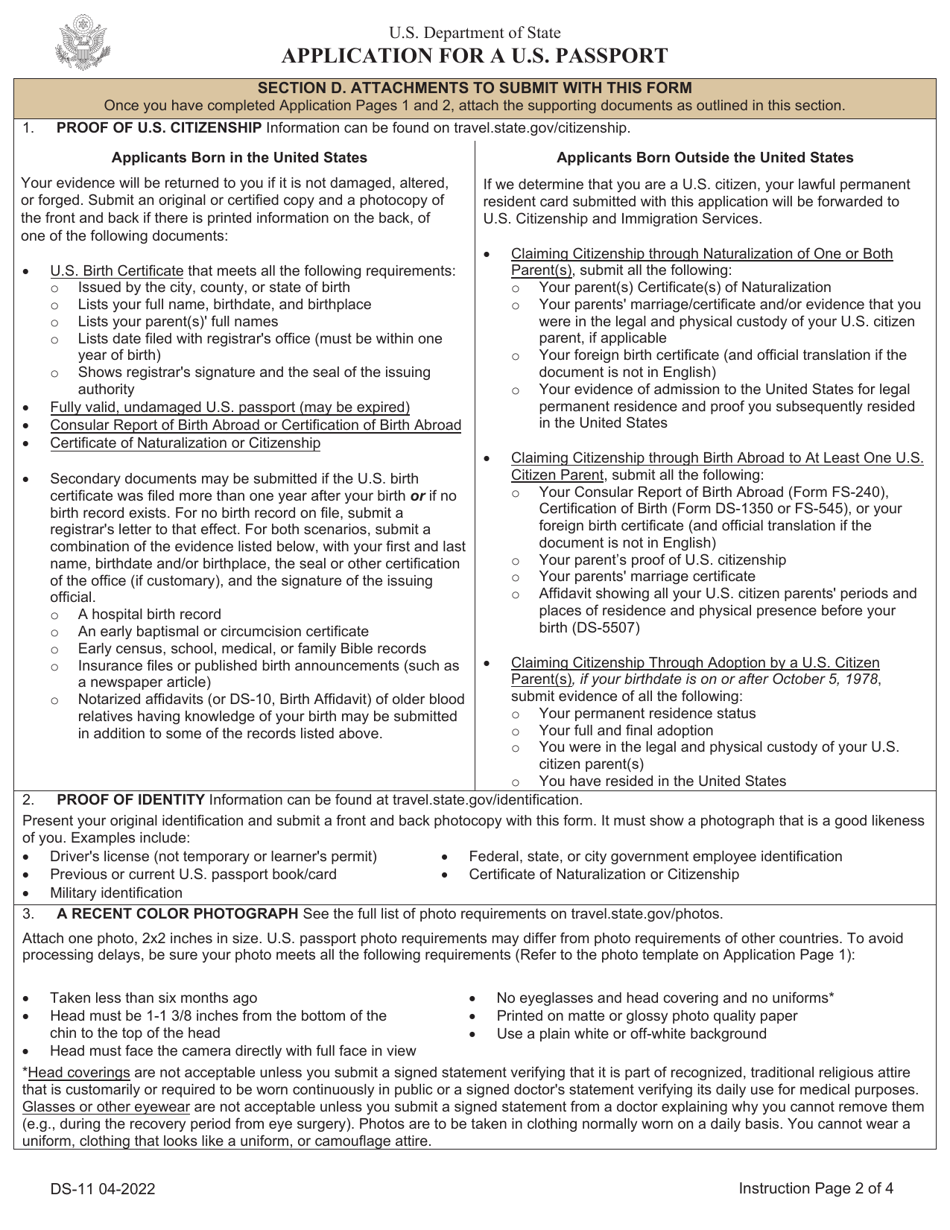
Form Ds 11 Download Printable Pdf Or Fill Online Application For A U S I am playing around with a prostaglandin molecule (23 heavy atoms) in gaussian09. my goal is to calculate the vertical ionization potential and electron affinity for the molecule. i already managed. Just few suggestions for finding a stationary point. you can add some additional angular flexibility into the basis by changing it to, say, 6 31g(2df,p) or even switch to more modern ahlrichs' def2 bases (def2svp to start from). besides, you'd better use ultrafine integration grid and tight optimization criteria and do geometry optimization and frequency calculation in a single run: opt=tight.