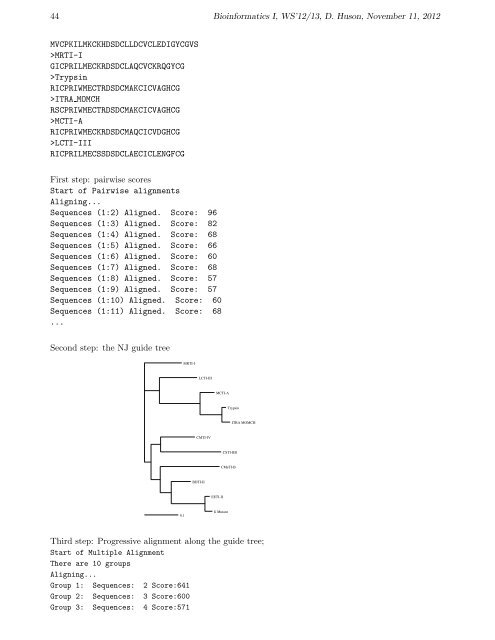
Github Sindhusita Sequence Alignment Algorithms Global Local And We explore deficiencies in existing multiple sequence global alignment algorithms and introduce a new indexing scheme to partition the dynamic programming algorithm hypercube scoring tensor over processors based on the dependency between partitions to be scored in parallel. We explore deficiencies in existing multiple sequence global alignment algorithms and introduce a new indexing scheme to partition the dynamic programming algorithm hypercube scoring.

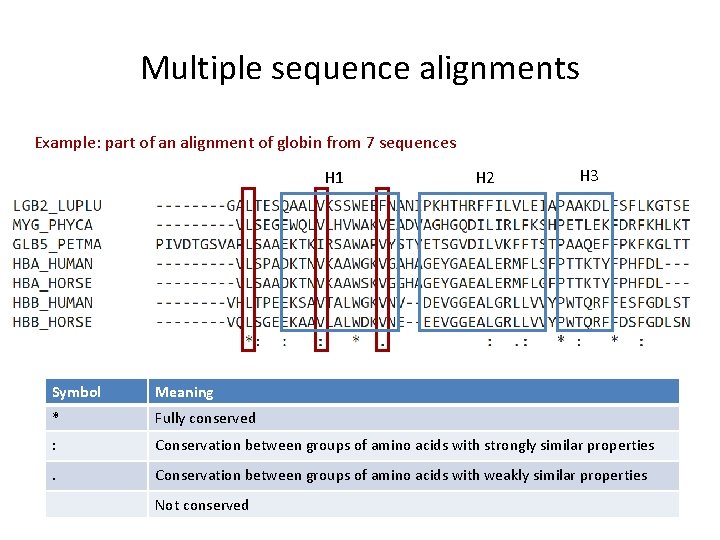
Multiple Sequence Alignment Algorithms For The Phylogenic Analysis A comparison of 10 most popular multiple sequence alignment (msa) tools, namely, muscle, mafft (l ins i), mafft (fft ns 2), t coffee, probcons, sate, clustal omega, kalign, multalin, and dialign tx is presented. In recent years improvements to existing programs and the introduction of new iterative algorithms have changed the state of the art in protein sequence alignment. this paper presents the first systematic study of the most commonly used alignment programs using balibase benchmark alignments as test cases. This paper provides a comprehensive comparative analysis of different multiple sequence alignment tools which are available today. the paper would first focus on different kinds of sequence alignment before moving to multiple sequence alignment, which then talks about the recent development in the algorithms and their techniques. Abstract in this paper, deep learning algorithms are compared for aligning multiple biological molecular sequences such as dna, rna, and protein. efficient algorithms are necessary for sequence alignment to identify significant insights, but there is a trade off between time and accuracy.
Bioinfor This paper provides a comprehensive comparative analysis of different multiple sequence alignment tools which are available today. the paper would first focus on different kinds of sequence alignment before moving to multiple sequence alignment, which then talks about the recent development in the algorithms and their techniques. Abstract in this paper, deep learning algorithms are compared for aligning multiple biological molecular sequences such as dna, rna, and protein. efficient algorithms are necessary for sequence alignment to identify significant insights, but there is a trade off between time and accuracy. We set forth to explore algorithms used for sequence comparison and their descendants applied to ngs assembly and alignment from a computational complexity perspective and in a comparative setting – where possible, in an effort to better understand their strengths and limitations. Accurate sequence alignments are essential for sequence comparison and for building structure activity models. high performance multiple sequence alignment (msa) algorithms have been suggested to improve the validity and relevance of in silico genomic experiments. A comparison of 10 most popular multiple sequence alignment (msa) tools, namely, muscle, mafft (l ins i), mafft (fft ns 2), t coffee, probcons, sate, clustal omega, kalign, multalin, and dialign tx is presented. In this review, the pairwise sequence alignment algorithms and the corresponding scoring system, heuristic algorithms for multiple sequence alignment and their defects, and quality estimation methods used to test multiple sequence alignment software are reviewed. there have been several reviews for multiple sequence alignment, such as refs [8, 9].
Overview Of Multiple Sequence Alignment Algorithms Yu He We set forth to explore algorithms used for sequence comparison and their descendants applied to ngs assembly and alignment from a computational complexity perspective and in a comparative setting – where possible, in an effort to better understand their strengths and limitations. Accurate sequence alignments are essential for sequence comparison and for building structure activity models. high performance multiple sequence alignment (msa) algorithms have been suggested to improve the validity and relevance of in silico genomic experiments. A comparison of 10 most popular multiple sequence alignment (msa) tools, namely, muscle, mafft (l ins i), mafft (fft ns 2), t coffee, probcons, sate, clustal omega, kalign, multalin, and dialign tx is presented. In this review, the pairwise sequence alignment algorithms and the corresponding scoring system, heuristic algorithms for multiple sequence alignment and their defects, and quality estimation methods used to test multiple sequence alignment software are reviewed. there have been several reviews for multiple sequence alignment, such as refs [8, 9].